About
To recognize and react against unknown proteins, B and T lymphocytes express unique, randomly generated receptors capable of evolving to refine their specificity. These receptors form the immune repertoire, a dynamic and highly diverse library, specific to each individual. The sequencing of these genes now makes it possible to map this repertoire, which provides information both on the individual's immunological history and on their future response capabilities.
But analyzing the repertoire remains a major challenge: each human being has 10^{11} unique receptors, the majority of which are exclusive to them. Above all, the interactions between these receptors and their target antigens are complex with current tools. In fact, the vast majority of antibody-antigen pairs are unknown, a veritable immunological “dark matter,” which our team is working to clarify by combining experimental and computational methods.
Experimental methods for measuring interactions : Our team develops high-throughput technologies, combining sequencing and molecular biology, to quantify the affinities between thousands of antibodies and their potential targets. These approaches make it possible to identify recognized antigens, discover antibodies of therapeutic or diagnostic interest, and study affinity maturation and antibody evolution.
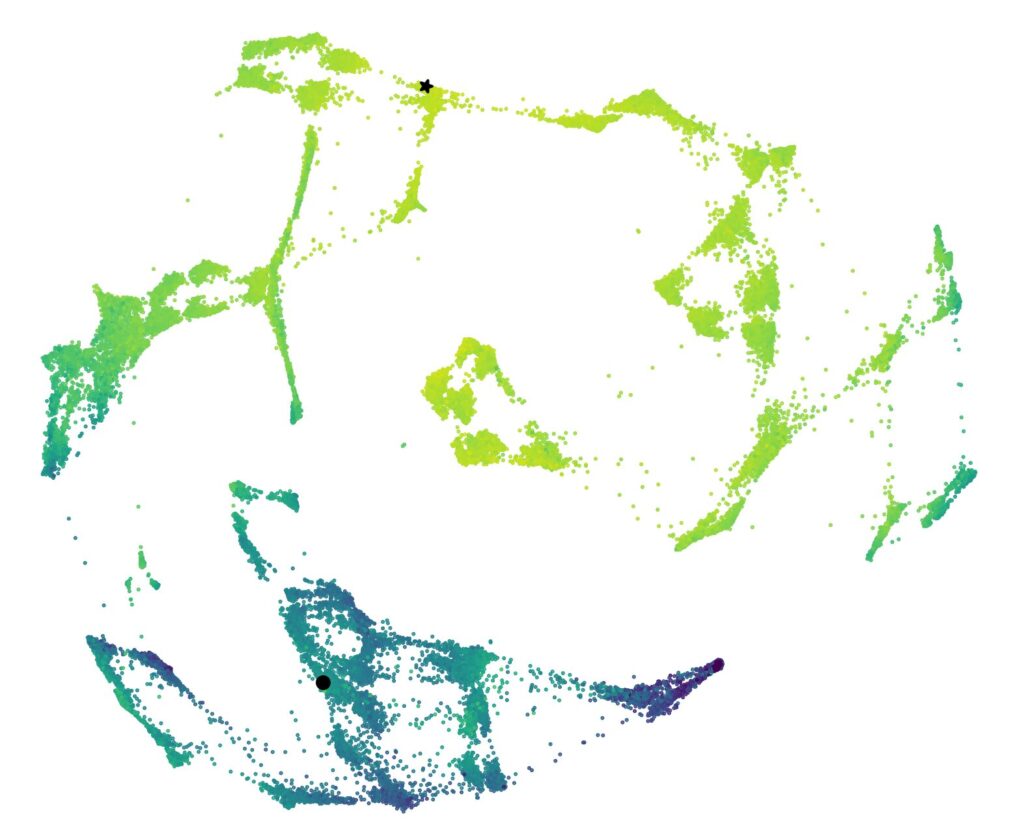
Computational models to decipher the repertoire In parallel, we design statistical models to analyze the diversity of repertoires, predict the receptors active against a given target, and simulate their evolution. By integrating experimental data and theories from physics and population genetics, we seek to understand the rules of immune recognition and generate virtual repertoires to test hypotheses.