Members















About
How do the immune system's sentinels translate the microbial and danger signals they receive into immunological instructions?
In the blood, mucous membranes, and lymphoid organs, dendritic cells play a dual role: they are both sentinels and conductors of the immune system. Hidden at the entry points used by pathogens, they locate infectious agents, ingest them, and release biochemical signals to alert the body's various lines of defense and attract antimicrobial cells to the site of infection. Once the intruder is digested, the dendritic cells expose fragments of the pathogen: the antigens, on their surface. They then migrate via the lymphatic pathways to secondary lymphoid organs (spleen, lymph nodes, mucosal-associated lymphoid tissues of the digestive tract and lungs), where they present these antigens to T and B lymphocytes. Once the characteristics of the pathogen have been identified, these cells, equipped with adapted and precise weaponry, in turn migrate to the site of infection to ensure its long-term eradication.
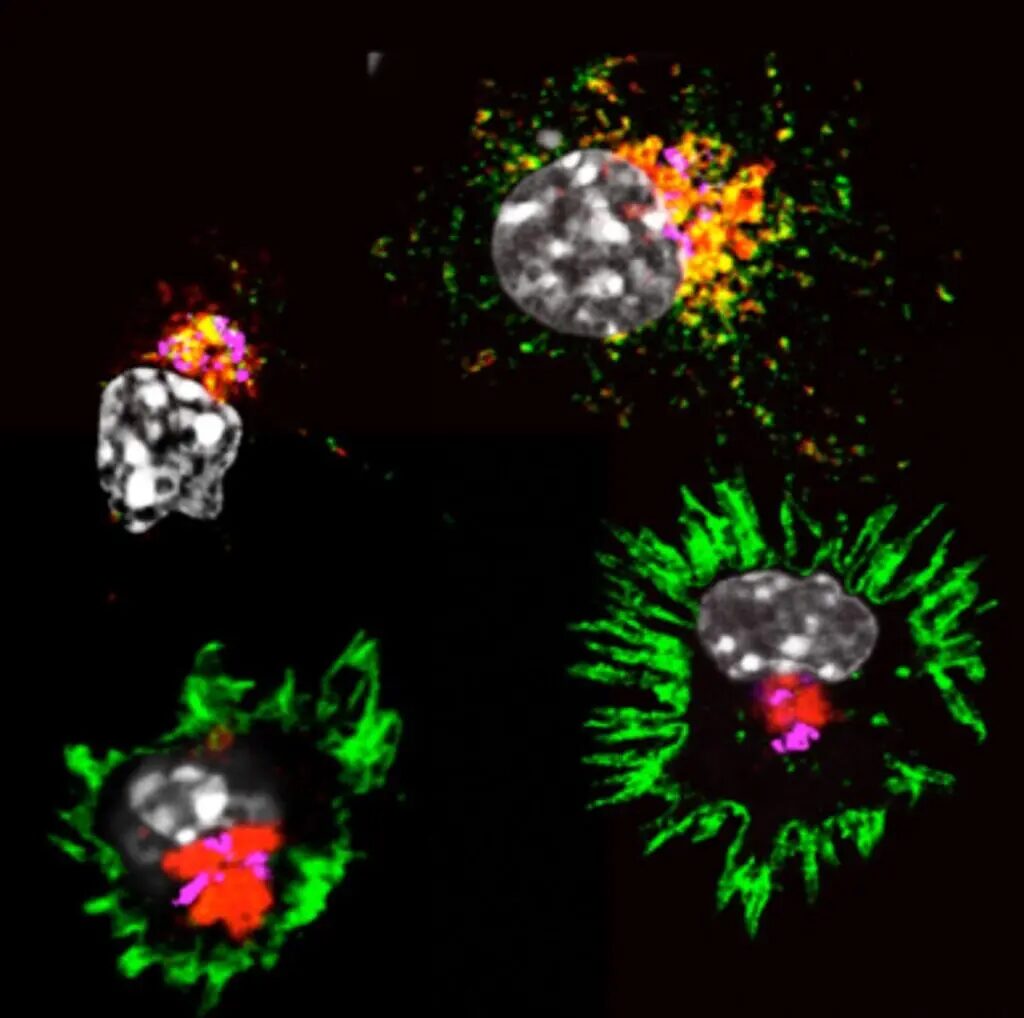
Confocal images showing dendritic cell activation over 24 hours (MHC II molecules in green, lysosomes in red, nuclei in gray). Image courtesy of Alexis Combes, CIML.
The Dendritic Cell Biology Laboratory (DeCiBeL) is committed to deepening understanding and developing innovative tools to study the cellular and molecular biology of dendritic cells (DCs) and other phagocytes, under normal and pathological conditions. Our research focuses on pathways involved in the identification of microbe-associated molecular patterns (MAMPs). These pathways orchestrate diverse cellular processes, including inflammation, autophagy, intracellular transport, endoplasmic reticulum (ER) stress, and energy metabolism, to efficiently activate immune cells. Autophagy and ER stress, particularly the misfolded protein response (UPR), are essential for regulating cellular metabolism and may link inflammation to environmental triggers in autoimmune diseases and cancer.
We study these pathways and the regulation of protein synthesis to better understand how these internal processes adapt DC biochemistry and metabolism to different microbial or danger environmental signals (MAMPs and DAMPs). The complex interconnections between these pathways require new high-resolution quantitative methods, to the development of which our laboratory regularly contributes. These innovations aim to deepen our understanding of how DCs acquire their distinctive immunoregulatory functions, with implications for clinical applications in cancer and autoimmune diseases.
Detailed information on the team's various ongoing projects can be found on the specific project pages, in addition to the brief description presented below.
Deciphering the interactions between ISR and PAMP detection.
P. Pierre (coordinator) with the support of all permanent researchers.
ER stress induces the misfolded protein response (UPR), via the activation of several sensors, including the endoplasmic reticulum (ER) kinase PERK (eIF2AK3), which, by phosphorylating its target eIF2a, reduces protein synthesis and establishes a biochemical program promoting stress resolution and cell survival, the integrated stress response (ISR). Our research has shown that DCs exhibit unusually high levels of phosphorylation of the translation factor eIF2a acquired during their cellular differentiation. eIF2a phosphorylation is mainly mediated by activated PERK activity in DCs, but without apparent signs of an active UPR (Mendes et al. 2022). This process has a low impact on protein synthesis levels and makes DCs relatively resistant to ER stress-inducing agents, such as the cytotoxin Subtilase (SubAB). We also showed that PERK regulates cell migration and is essential for responding to MAMP detection. Surprisingly, PERK stimulation by SubAB induces type-I interferon expression in plasmacytoid DCs (pDCs), which may also act synergistically with Toll-like receptor 7 stimulation (Barros et al. 2025). IFN induction in response to SubAB alone relies on STimulator of Interferon Genes (STING) activation coupled with PERK-mediated inhibition of protein synthesis. By blocking mRNA translation via the ISR, SubAB precipitates the downregulation of negative feedback regulators of innate signaling, required to release TBK1 signaling associated with STING activation.
These results, along with the identification of mutations in other eIF2AK genes in families of patients susceptible to autoimmunity or autoinflammation, highlight the importance of the ISR in human autoimmunity. In particular, the discovery of a SAVI patient with mutations in the Sting1 and Perk genes and displaying symptoms unusual for this interferonopathy (de Cevin et al. 2023), highlights the relevance of better understanding the crosstalk between the ISR and innate immunity.
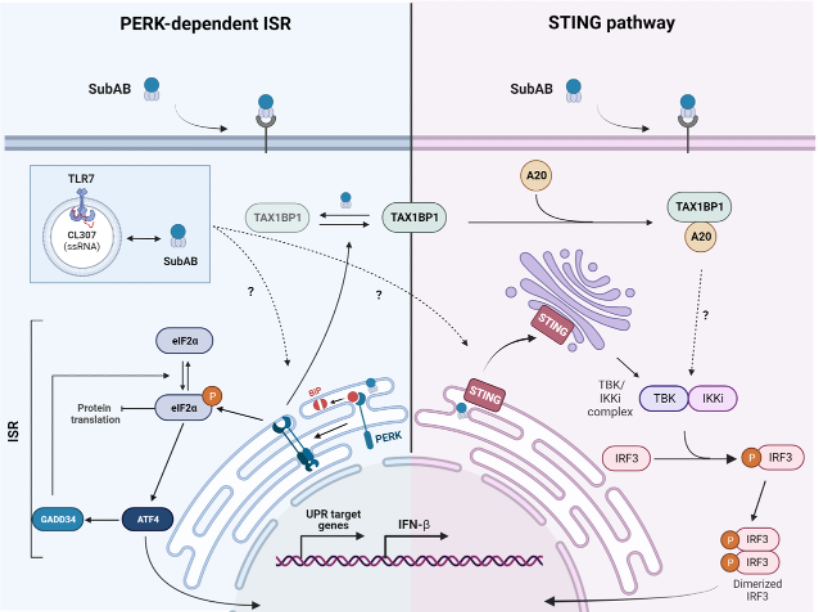
SubAB induces type I IFN production via STING activation and loss of the negative feedback regulator TAX-1BP1, which is decreased upon protein synthesis shutdown.
To facilitate our studies, we established a web-accessible database (MetamORF, https://metamorf.hb.univ-amu.fr) to identify uORF/ISR-regulated genes (in collaboration with C. Brun, TAGC Marseille, Choteau et al. 2022). We also developed a novel flow cytometry-based method (SNUPR, co-coordination with RJ. Argüello) to simultaneously monitor the activation of the three UPR branches. SNUPR has proven to be a valuable tool to detect the importance of the IRE-1 branch in the acquisition of Bortezomib resistance by multiple myeloma cancer cells (Gigan et al. 2024).
Endomembrane dynamics in innate and adaptive immune sensing and signaling.
E. Gatti (coordinator), with the support of all permanent researchers.
MAMP sensing impacts the organization of membrane organelles, and in particular endocytosis, to enable dendritic cells to efficiently acquire their immunostimulatory functions. We have shown that the BAD-LAMP/LAMP5 protein is mainly present in human pDCs and pDC blastic neoplasms (BPDCN), and controls the transport of TLR9 to signaling endosomes (Combes et al., Nat Com. 2017). Our results also indicate that BAD-LAMP is expressed in patient-derived MML leukemia cells, where it maintains cell survival and cancer progression via activation of NF-kB signaling (Maldonado et al. 2022).
In parallel, we are continuing to study the role of RUFY proteins in phagocytes (Char et al. 2020). The RUFY family is characterized by a RUN domain, which interacts with small GTP-binding proteins, and a FYVE domain, involved in the recognition of PtdIns(3)P lipids in organelles. In addition to the regulation of autophagy in DCs, our study suggests a role for RUFY4 in the selective elimination of mitochondria in alveolar macrophages via effectors such as PLEKHM1, Rab7, the HOPS complex, and various mitochondrial proteins (Valecka et al. 2022). We also demonstrated that an immune cell-specific isoform of RUFY3 participates in lysosome clustering in macrophages and DCs in response to bacterial LPS (Char et al. 2023). Overall, RUFY3 plays a regulatory role in IFN-g signaling, antigen presentation, and migration, and possesses an anti-inflammatory role in vivo. RUFY3 interacts with the small GTPase ARL8b, which controls dynein-mediated transport of late endosome along microtubules in relation to the formation of PtdIns(3)P lipids by the class III PI3 kinase, Vps34. In a parallel project, we also confirmed that pharmacological inhibition of VPS34 interferes with RUFY3 distribution, but also with TLR7/9 signaling.
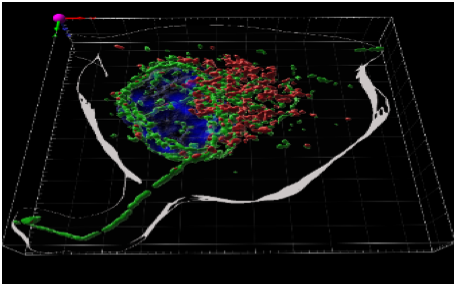
Confocal image showing the distribution of RUFY3 (green) on tubules emanating from late endosomes (Lamp1, red) in a situation where PtdIns(3)P accumulates in macrophages. Image courtesy of Rémy Char, CIML
Metabolic Regulation of Immunity.
Rafael J. Argüello (Coordinator)
We aim to elucidate how metabolic processes regulate immune responses in human myeloid cells, including DCs, macrophages, and monocytes, in the contexts of cancer and sepsis. Our laboratory has developed SCENITH, a pioneering flow cytometry-based method enabling metabolic profiling at the cellular level. This innovation, detailed in our publication (Argüello* et al., Cell Metabolism, 2020), has become a cornerstone for metabolic studies of immune cells, accumulating over 400 citations. Other methodological advances include SunRiSE, assessing single-cell translation elongation rates, and recently SNUPR (Gigan et al. 2024), enabling single-nuclear analyses of the response to misfolded proteins. These tools have significantly improved our understanding of metabolic and proteostatic regulation in immune cells, particularly highlighting metabolic changes associated with disease states. We recently introduced an adaptation of SCENITH suitable for fixed and stabilized blood samples, its compatibility with sorting and epigenomic analysis, thus expanding its application for future clinical studies. Our studies have also revealed metabolic vulnerabilities in leukemia, highlighting how mitochondrial adaptations contribute to resistance to therapies (Bosc et al., 2021). Other findings have identified metabolic states governing the differentiation of inflammatory and tolerant dendritic cells, influencing immune modulation and patient outcomes (Adamik et al., Nat Commun, 2022).
Dendritic cell response to modified vaccine mRNAs.
Béatrice Nal-Rogier (Coordinator) with the support of all permanent researchers.
Description: The impact of finely tailored mRNAs on research and health science is expected to be immense. The design and rapid market introduction of the SARS-CoV-2 Spike mRNA vaccine during the last pandemic years illustrated the novelty and power of the approach, while highlighting correlated improvements aimed, for example, at better tolerability/side effects and improved storage at room temperature. We are investigating the impact of mRNA methylations by novel AI-designed methyltransferases on mRNA stability, protein translation, and molecular pathways in human DCs. Our partners at AFMB and IGF will design and produce novel mRNA methyltransferases, which will be evaluated in the laboratory for their translation efficiency and induction of innate immune responses (IFN-I/III, pro-inflammatory cytokines), the integrated stress response (ISR), and antigen presentation.
The DeCiBeL laboratory.
Through our work and the establishment of numerous international collaborations, our team strives to better understand how each of the pathways described above functions and interacts to lead to an effective immune response. We also aim to identify the main effector molecules, so that we can manipulate these pathways using targeted pharmacology or corrective genetics. The goal of these interventions is to reduce the hyperactivity of the immune system that causes autoimmune diseases or, conversely, to improve the effectiveness of DC stimulation to elicit a potent immune response against infectious diseases and cancer.
Projects
Project: UNFIRE Other participants: Ciphe (Ana Zarubica), Hospices Civils de Lyon and CIRI (Sophie Trouillet-Assant, Thierry Walzer), CoVir team […]





The integrated stress response (ISR) is a cellular pathway activated by various biochemical triggers. It reduces global protein synthesis while […]

Project: IMPACT Sensors Other participants: IGBMC, Strasbourg (Nicolas Charlet-Berguerand) and Institut Curie, Paris (Franck Perez). Duration 2025 – […]

Partner: University of Milan, Italy (Prof. Angelo Poletti, DISFEB) Frameshift mutations in HSPB8 cause neuromyopathies by disrupting […]

CNRS – University of Melbourne Graduate Research Projects The project investigates the molecular interplay between the E3-ubiquitin ligase MARCH1 and […]

Duration: 2021–2025.Partners & funding amounts: APHM/La Timone (E. Tabouret); amount for team: 325 k€.Responsibilities: PI–coordinator (only beneficiary)Theme & content of […]

Bone marrow metabolic niches regulate hematopoiesis (MetaNiche, ANR-22-CE15-0015-02). Duration: 2023–2027 (42 months).Partners & funding amounts: Institut Curie (L. Perié), Institut […]

Full title & acronym: Biomarkers established to stratify sepsis long-term adverse effects to improve patients' survivorship (BEATsep).Duration: 2024–2029 (60 months).Partners […]

Full title & acronym: Understanding lipid immunometabolism to treat disease (UNLIMITED).Duration: 2025–2029 (48 months).Partners & funding amounts: Consortium led by […]

Anti-GD3 CAR-NK cells to overcome therapeutic resistance in glioblastoma (Caribbean). Type: National.Duration: 2025–2029 (48 months).Partners & funding amounts: INP-APHM (E. […]

Targeting intratumor metabolic heterogeneity in neuroblastoma to develop innovative treatments (IMAGINe) Type: National.Duration: 2024–2029 (48 months).Partners & funding amounts: CRCM […]

Targeting acute myeloid leukemia immunosuppressive microenvironment by combined IDO1 inhibition and PD-1 blockade Type: European.Duration: 2023–2027 (36 months).Partners & funding […]

South-Research On Cancer for Kids. INCA Pediacriex (France) Type: National (integrated cancer site).Full title & acronym: South-Research On Cancer for […]

Tailoring CAR T-cell metabolism and activation to enhance therapeutic efficacy (CARma) Type: National.Duration: 2025–2029.Partners & funding amounts: CRCM (R. Lasserre); […]