

Facility: Computational Biology, Biostatistics & Modeling (CB2M)
About
CB2M (Computational Biology, Biostatistics & Modeling) is CIML's computational biology platform dedicated to advancing immunology research at CIML through computational biology. It offers expertise in several areas, including the development of computational methods and tools, biological data analysis, statistics, and physical and mathematical modeling.
By creating interdisciplinary links between immunologists and computational biology disciplines, CB2M aims to bring the most innovative approaches in computational biology to CIML.
To address these complex challenges, CB2M operates according to two complementary strategic approaches:
1. A group of expert scientists
Computational biology has become a fundamental pillar of contemporary immunological research. As technology advances, data analysis methods are becoming increasingly sophisticated, incorporating mathematical and statistical concepts of unprecedented complexity. These algorithmic approaches now make it possible to explore previously inaccessible immune dimensions, transforming our understanding of the immune system. However, using modern computational tools for biology requires a unique combination of skills.
CB2M scientists bring these skills to CIML:
- in data analysis and statistics, thanks to their expertise in bioinformatics and pure and applied mathematics;
- in mathematical and physical modeling of natural systems, thanks to their initial training in theoretical physics;
- in theoretical computer science and software engineering, thanks to their training and professional experience;
- in interdisciplinary relations, thanks to their past interactions with many biologists.
2. A cooperative ecosystem
Research projects conducted at CIML require in-depth, complex, and time-consuming computational biological analyses. These analyses rely on increasingly advanced technologies (e.g., single-cell omics, spatial omics) that are increasingly combined with cytometry, imaging, and histology. CB2M is committed to helping researchers meet these challenges and enable them to produce scientific work of the highest quality.
Recruiting one or more bioinformaticians specifically dedicated to their projects is the most efficient way for CIML teams to perform robust computational biology analyses. However, for most principal investigators in biology, recruiting and supervising bioinformaticians is a challenge. Furthermore, isolated bioinformaticians lack opportunities to interact with experts in their field to adopt best analytical practices.
At CIML, CB2M provides a collaborative environment where biologists and bioinformaticians can easily interact. Here, bioinformaticians are surrounded by colleagues in their field and supported by experts, but they maintain close ties with their original team. Within its collaborative ecosystem, CB2M provides expertise, training, and mentoring to all CIML bioinformaticians.
Training
CB2M is committed to training CIML staff in the knowledge, methods, and techniques associated with computational biology and statistics. CB2M has developed a comprehensive training program for bioinformaticians and biologists wishing to acquire further knowledge and skills in computational biology.
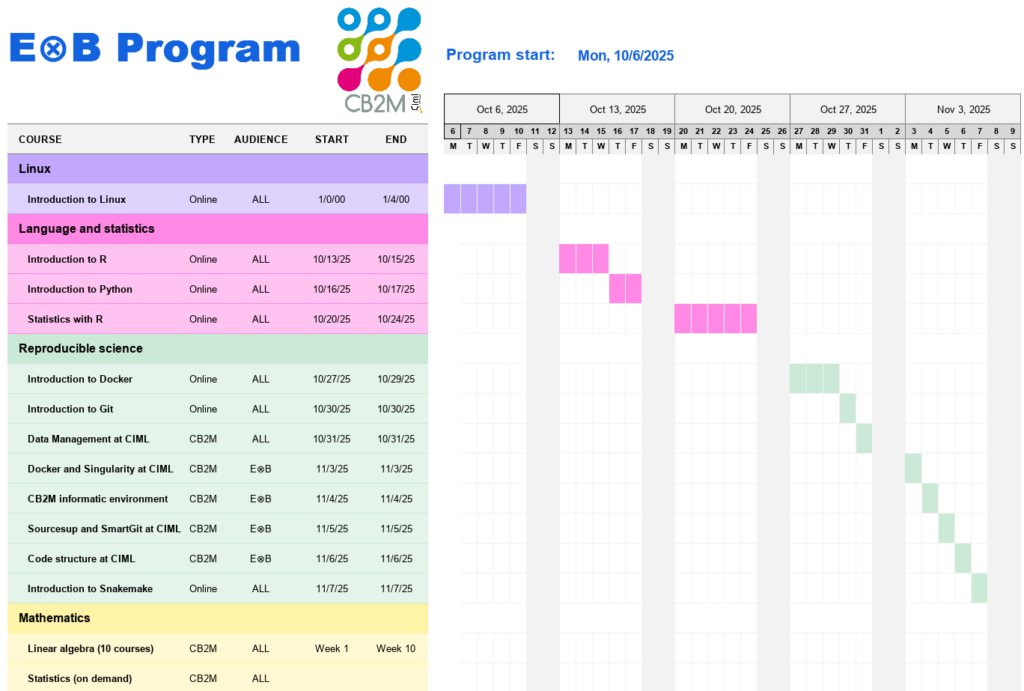
This program, called Program E⊗B (Experimentalist Cross Bioinformatician Program), includes the following modules (module details are shown in the Gantt chart below):
- Linux: This module aims to provide basic knowledge and practices on the Linux operating system.
- Language and Statistics: This module introduces the R and Python languages as well as the application of basic statistics for data analysis.
- Reproducible Science: This module offers courses aimed at providing methodologies, tools, and best practices to ensure the reproducibility of bioinformatics analyses. This area is the common core of all courses.
- Mathematics: this module aims to teach or deepen the mathematical knowledge most commonly used in analyses carried out at CIML.
The program is tailored to the needs of each individual:
- For bioinformaticians (interns, doctoral students, engineers, post-docs), the courses are compulsory and are selected according to the learner's level.
- For biologists who wish to acquire dual skills in biology and bioinformatics (mainly doctoral students), it is proposed to follow the complete E⊗B program with a schedule adapted to their objectives.
Please note that in addition to the E⊗B program:
- CB2M offers courses on specific topics for CIML staff, such as single-cell RNA-seq data analysis using the ShIVA interface or statistics. These courses are announced well in advance and are subject to voluntary registration.
- CB2M members also provide training for external courses, such as Inserm training. CIML members are invited to register for these courses if the subject interests them.
Here is an example of a schedule for the E⊗B Program:
Open Science
The CB2M is strongly committed to open science and scientific reproducibility. In order to guarantee the reliability and transparency of research results, the CB2M has implemented good practices and methods to ensure the 100 % reproducibility of all bioinformatics analyses carried out within the framework of scientific projects.
A key element of this mission is the training : each bioinformatician recruited at CIML, as well as the students welcomed for internships, is trained in these practices and supported to ensure their proper application. This approach contributes to establishing a true culture of rigor and reproducibility in all of CIML's computational biology activities.
The CB2M also plays an active role in the adoption of national infrastructures for open science. It thus supports the use of Research Data Gov, the official French platform for storing scientific data. Thanks to CB2M's involvement, CIML is one of the first Aix-Marseille University laboratories to have its own dedicated space within the Data Gouv Research warehouse. CB2M also participates in the National Conference on Research Data (ANDOR).
Through these actions, the CB2M strengthens the openness, transparency and long-term impact of immunology research carried out at the CIML.
Projects
Project: DECITIP Plasmacytoid dendritic cells (pDCs) excel in the production of type I and III interferons (IFN), […]


Our health depends on a delicate balance between immune defense against pathogens and tolerance to […]


In collaboration with Denis Puthier (TAGC) and Lionel Spinelli (CB2M, CIML), we co-developed SciGeneX, an innovative R package allowing […]

Lymph nodes are formed in the embryo by Lymphoid Tissue Inducer (LTi) cells. LTi cells arise not from the yolk […]

Innate immune cells are essential partners of the central nervous system, supporting both homeostasis and repair. Within the brain, perivascular […]

Project: Treating Breast Cancer Breast cancer is the most frequently diagnosed cancer in women in […]

Project: TREATLIVMETS Liver metastases occur in nearly 50% of cancer patients, particularly in […]

Project: pDCComp Individuals infected with viruses or suffering from inflammatory diseases develop a set of behavioral disorders […]

Project: PLBIO 2024 Coordinator: Thierry Walzer, CIRI, Lyon. Other participant: Nathalie Bendriss-Vermare, CRCL, Lyon. Abstract: Natural Killer cells […]

Project: ACLAMADABREC Partly through their effectiveness in activating natural killer (NK) cells and CD8 T lymphocytes […]

Project: MITI Other participants: Magali Richard, LIG, Grenoble; Lionel Spinelli, CB2M hub of CIML. Summary: This project aims to […]

Project: UNFIRE Other participants: Ciphe (Ana Zarubica), Hospices Civils de Lyon and CIRI (Sophie Trouillet-Assant, Thierry Walzer), CoVir team […]





Project: PLBIO-25 Coordinator: Marianne BURBAGE, Institut Curie, Paris. Other participant: Andrew GRIFFITHS, ESPCI, Paris. The effectiveness of CD8 T lymphocytes […]

Project: DCFUNDEVSCREEN Abstract: Studies in mice have shown the key antiviral/tumor role of plasmacytoid dendritic cells (pDCs) […]

Project: RIPIREVI Other participants: CIPHE (Ana Zarubica and colleagues). Abstract: Type I interferons (IFN-Is) are essential for […]


Project: LYMPHOMATLAS Single-cell multi-omic studies in B-lymphomas have highlighted a […]


Project: NRegSkin Project leader: Mei LI (IGBMC, Strasbourg). Other participants: Katia Boniface, Julien Seneschal (University of Bordeaux). Summary: The skin […]

The progression of follicular lymphoma is marked by iterative cycles of remission and relapse, the latter usually resulting from the […]

Project: REVOLUTION Coordinator: Edouard Sage, Foch Hospital, Suresnes. Other partners: Isabelle Schwartz-Cornil, INRAE, Jouy-en-Josas; Jérémie Pourchez, Ecole des Mines […]

Project: ICARO Current acellular vaccines are mostly effective against extracellular pathogens, but fail to induce a response […]


Project: DeCoDis In the context of a European consortium, we are investigating the dynamics and function of border-associated macrophages […]

In order to identify new therapeutic combinations and understand the mechanisms of resistance to targeted therapies in lymphoid-derived hemopathies […]

Patients with follicular lymphoma (FL) present a very high biological heterogeneity, which strongly impacts the response to current therapies. […]

We have demonstrated that the neurokine TAFA4, produced by sensory neurons in the skin, can promote tissue repair […]

The host response to pathogens is orchestrated by the immune system and the nervous system, both present in […]

Beyond the pain perception processes that are induced in the event of injury or infection of the […]

The TCELL-CODE project, coordinated by Dr. Rémy Lasserre at the Marseille Cancer Research Center, aims to elucidate […]


The structuring action Spatial Omics of the CALYM consortium, coordinated by Dr Pierre Milpied, aims to develop a virtual platform dedicated […]

The GCselection project, coordinated by Dr. Pierre Milpied (CIML), revisits the mechanisms of antibody maturation in centers […]


The ST-omics structuring action of the Cancéropôle de la Région Sud is co-coordinated by Dr Pierre Milpied (CIML), Prof Emmanuelle […]

The AITLas-Impact project, coordinated by Pierre Milpied (CIML) in collaboration with Professor François Lemonnier (Mondor Institute of Biomedical Research, […]

Dr. Pierre Milpied's team at CIML is participating in a collaborative project coordinated by Dr. Jean-Pierre de Villartay (Institut […]


