Publication: FB5P-seq: FACS-Based 5-Prime End Single-Cell RNA-seq for Integrative Analysis of Transcriptome and Antigen Receptor Repertoire in B and T Cells.
Publié dans: Front Immunol 2020 ; 11(): 216
Auteurs: Attaf N, Cervera-Marzal I, Dong C, Gil L, Renand A, Spinelli L, Milpied P
Résumé
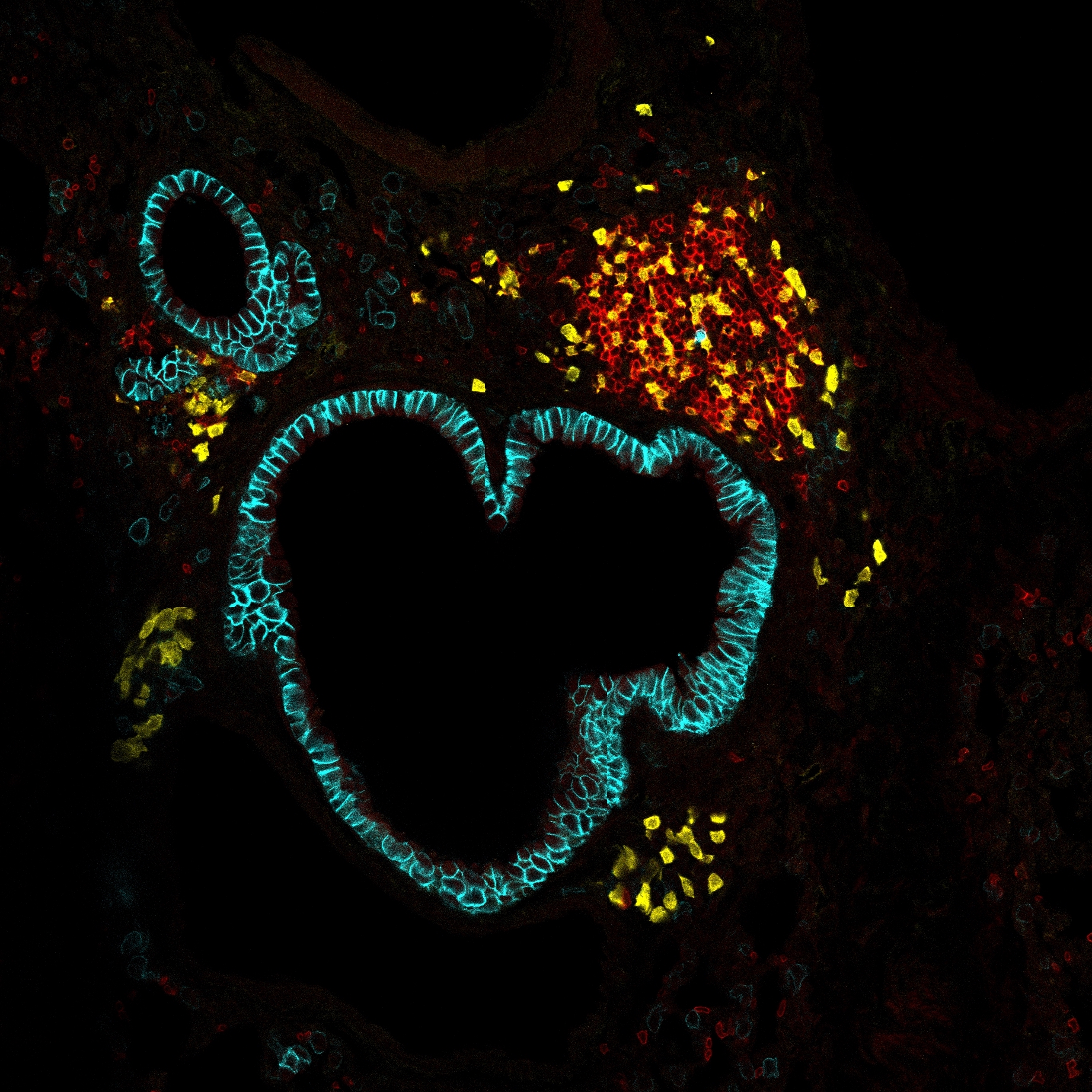
Single-cell RNA sequencing (scRNA-seq) allows the identification, characterization, and quantification of cell types in a tissue. When focused on B and T cells of the adaptive immune system, scRNA-seq carries the potential to track the clonal lineage of each analyzed cell through the unique rearranged sequence of its antigen receptor (BCR or TCR, respectively) and link it to the functional state inferred from transcriptome analysis. Here we introduce FB5P-seq, a FACS-based 5′-end scRNA-seq method for cost-effective, integrative analysis of transcriptome and paired BCR or TCR repertoire in phenotypically defined B and T cell subsets. We describe in detail the experimental workflow and provide a robust bioinformatics pipeline for computing gene count matrices and reconstructing repertoire sequences from FB5P-seq data. We further present two applications of FB5P-seq for the analysis of human tonsil B cell subsets and peripheral blood antigen-specific CD4 T cells. We believe that our novel integrative scRNA-seq method will be a valuable option to study rare adaptive immune cell subsets in immunology research.
Lien vers Pubmed [PMID] – 32194545
Lien vers HAL – inserm-02561110
Lien vers le DOI – 10.3389/fimmu.2020.00216