Publication: SciGeneX: enhancing transcriptional analysis through gene module detection in single-cell and spatial transcriptomics data.
Publié dans: NAR Genom Bioinform 2025 Jun; 7(2): lqaf043
Auteurs: Bavais J, Chevallier J, Spinelli L, van de Pavert SA, Puthier D
Résumé
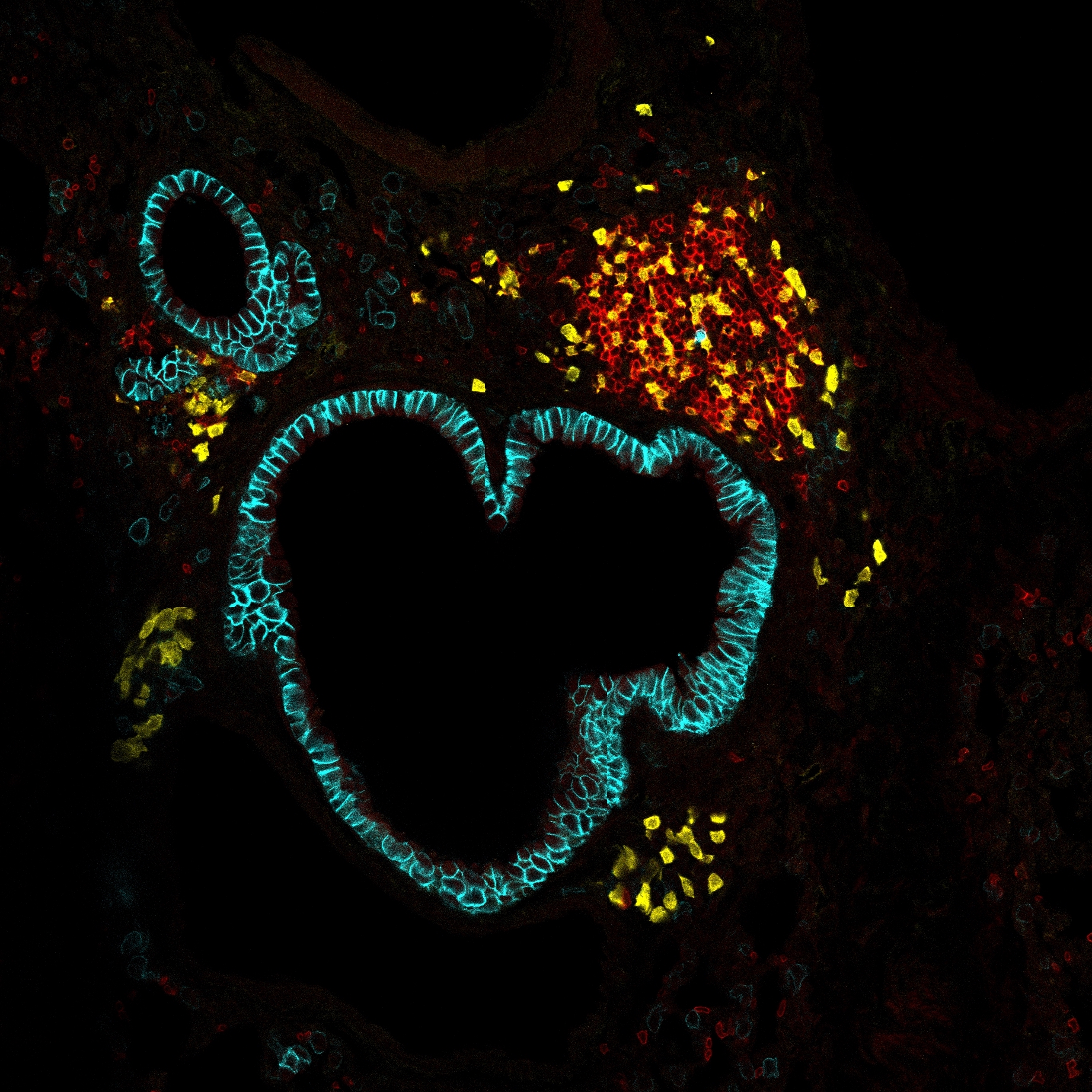
The standard pipeline to analyze single-cell RNA-seq or spatial transcriptomics data focuses on a gene-centric approach that overlooks the collective behavior of genes. However, understanding cell populations necessitates recognizing intricate combinations of activated and repressed pathways. Therefore, a broader view of gene behavior offers more accurate insights into cellular heterogeneity in single-cell or spatial transcriptomics data. Here, we describe SciGeneX (Single-cell informative Gene eXplorer), a R package implementing a neighborhood analysis and a graph partitioning method to generate co-expression gene modules. These modules, whether shared or restricted to cell populations, collectively reflect cellular heterogeneity. Their combinations are able to highlight specific cell populations, even rare ones. SciGeneX uncovers rare and novel cell populations that were not observed before in human thymus spatial transcriptomics data. We show that SciGeneX outperforms existing methods on both artificial and experimental datasets. Overall, SciGeneX will aid in unravelling cellular and molecular diversity in single-cell and spatial transcriptomics studies.
Lien vers Pubmed [PMID] – 40248490
Lien vers le DOI – 10.1093/nargab/lqaf043